
|
RigidFinder: rigid blocks derived from comparison of protein
structure conformations |
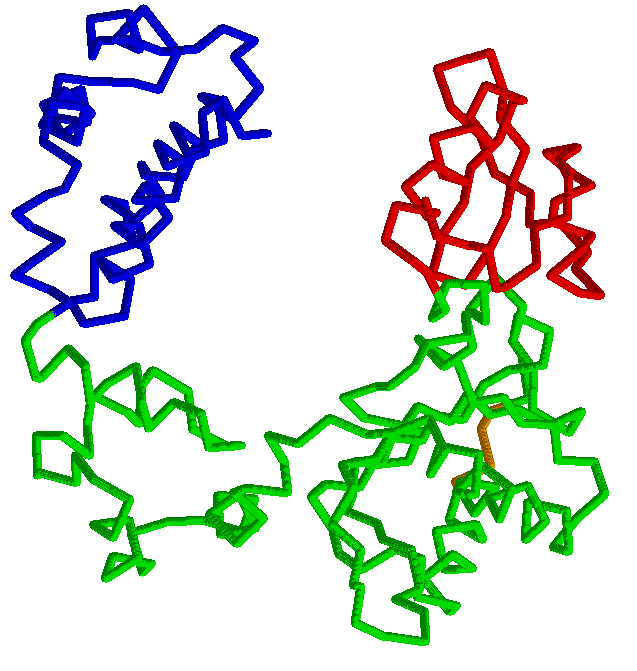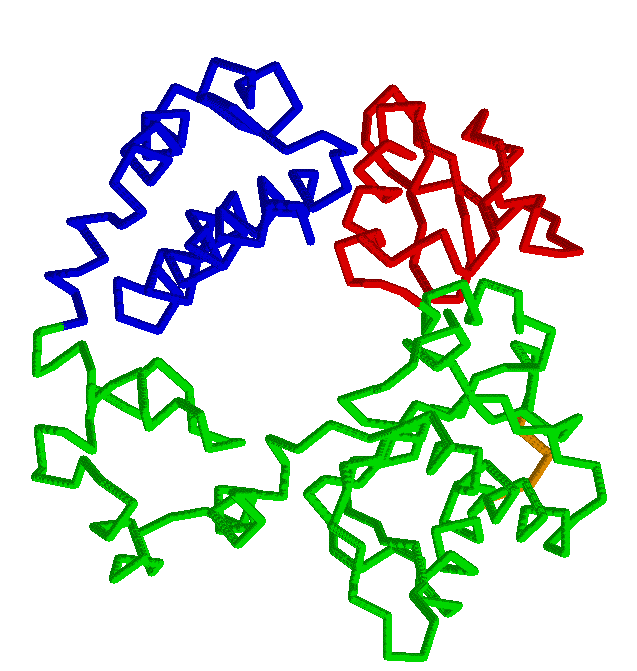
Comparison of two protein conformations with the aim of identifying movable rigid regions is a crucial part of protein motion analysis. Advances in crystallography and new methods of structure determination enabled crystallization and analysis of large macromolecular complexes, such as RNA polymerase II or GroEL, often in different conformations. The availability of several conformations for macromolecular multichain complexes with ten thousands of residues poses a need for new analysis methods and tools capable of extracting useful information from the different conformations. RigidFinder is a novel method for identification of protein rigid blocks from two different conformations. Rigid blocks are defined as blocks where all inter residue distances are conserved across conformations. Distance conservation, unlike averaged values, e.g. RMSD, used by absolute majority of other method, allows for sensitive identification of rigid blocks, with RigidFinder capable of detecting rigid blocks in the full range of sizes from large subunits to loops as small as 4 residues long. Distinguished feature of the method is that it is capable of finding rigid blocks that consist of non-consecutive fragments of one or multiple polypeptide chains. An efficient quasi-dynamic programming search algorithm utilized by the method allows for its real time application to analysis of large macromolecular complexes. The method is integrated in software Belka and can be freely downloaded.Abyzov A, Bjornson R, Felipe M, Gerstein M. RigidFinder: a fast and sensitive method to detect rigid blocks in large macromolecular complexes. Proteins. 2010 Feb 1;78(2):309-24. |